Controlled indexing for question-answering in a specialized domain
Thierry Delbecque and Pierre Zweigenbaum
Contents |
Abstract
Indexing named entities is useful for answering open domain questions. In the same way, recognizing and indexing specialized (here, medical) named entities in documents is important for question answering in the medical domain. This raises specific issues which we first describe. We then report on two lines of work conducted in this context.
Issues raised by medical named entity recognition
Controlled vocabulary terms vs Named entities
Named entities comprise proper nouns (names of persons, organizations, geographic locations, etc.) and numeric expressions (dates, monetary amounts, etc.). Proper nouns are capitalized or include specific markers (President, Incorporated, etc.). What we call here medical named entities are actually controlled vocabulary terms, i.e., terms which are all known in advance and listed in a reference thesaurus. The main problem is that these terms can occur under variant forms. This problem is made all the more acute as the number of medical terms is quite large---the UMLS Metathesaurus (version 2007AC) contains 1.4 million concepts and 5.3 million different terms; it includes over 100 biomedical terminologies, among which the MeSH thesaurus, the International Classification of Diseases (ICD-9 and ICD-10), and the SNOMED Nomenclature (see Lindberg et al., 1993).
Controlled indexing: mapping to reference concepts
The controlled vocabulary term recognition task, or controlled indexing task, includes the recognition of terms and their variants and the identification of the associated concepts. Here, a concept is represented by a unique identifier. A concept has an associated preferred term and may also have additional synonym terms. The goal of controlled indexing is to find the concept identifiers associated to terms or term variants which occur in a given input text. it can be seen as a (multilabel) categorization task (aka supervised classification): assigning one or more categories (concept identifiers) to an input text.
The two experiments reported below take each of these two views of controlled indexing: term recognition, then multilabel categorization.
Efficient recognition of UMLS Metathesaurus terms in scientific articles
Context
The reference system for recognizing medical entities in English text is MetaMap Transfer (MMTx) (Aronson, 2001). MMTx identifies terms of the UMLS Metathesaurus in running text, taking into account diverse morphological, syntactic and synonymic variants of the initial UMLS terms. However, preliminary experiments using this indexer led to too long computing times and too much memory usage. It motivated us to write a lightweight UMLS tagger (MetaCoDe), that could run using less memory and proceed in a shorter time (Delbecque & Zweigenbaum, 2007). The saved time could then be devoted to specific QA-oriented computations.
The main time-consuming factors that we identified in MMTx are the following:
- MMTx processes a text using the full set of concepts and terms present in the UMLS Metathesaurus, which causes heavy disk access;
- MMTx tests numerous variants of the input words, using the UMLS SPECIALIST knowledge bases and tools: acronyms and abbreviations, synonyms, and derived words.
Methods
To obtain faster processing, we used two principles:
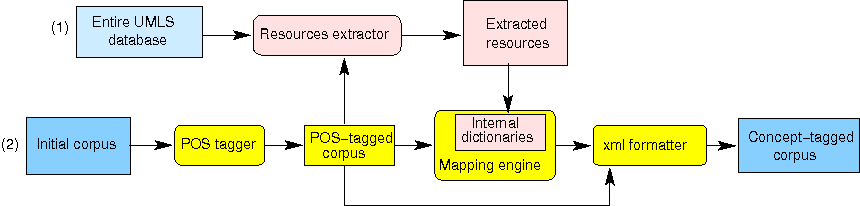
- Adopt a two-pass strategy (see Figure 1):
- The first pass projects the UMLS Metathesaurus on the corpus: it builds a reduced subset of UMLS resources which is sufficient to further process the corpus.
- The second pass is the actual mapping algorithm, which relies on the reduced subset prepared in the first pass instead of the whole UMLS. The reduction in data size enables the program to drastically reduce disk access.
- Trade some of the sophistication of MMTx for higher efficiency:
- Term variation in MetaCoDe is limited to explicit synonyms present in the Metathesaurus and to inflected forms.
- Abbreviation and derivation, if not present in the Metathesaurus, are not addressed.
A schema of MetaCoDe, with its two passes, is shown on Figure 1.
Evaluation
Two questions are raised by this work:
- Is the gain in speed substantial?
- What is the quality of MetaCoDe relatively to MMTx?
Speed was evaluated on a corpus of 7,260 medical abstracts extracted from MEDLINE, totalling 2,160,613 words, using the same machine (Pentium 4 biprocessor, 3GHz, 1.5 Go; no other task running). MetaCode ran in 26 minutes, obtaining a 100x gain in speed over MMTx (50 hours).
Mapping quality was evaluated on an independent random sample of 30 medical MEDLINE abstracts, totalling about 9,200 words. MMTx results for this corpus were taken as the gold standard against which MetaCoDe output was compared. Table 1 shows precision, recall and F-measure (harmonic mean of P & R) of MetaCode. The decrease in recall is significant: MetaMap, but not MetaCode, could detect some concepts under variant forms that were not listed in the UMLS. Nevertheless, it is not as dramatic as one might have expected. Furthermore, easy improvements of the algorithm can be achieved by simply modifiying the pattern set used to build noun phrases, e.g., by including isolated verbal forms as MetaMap does.
| Precision | Recall | F-measure | Speed |
|---|---|---|---|
| 0.93 | 0.76 | 0.83 | 100x |
Indexing ICD-9-CM diagnoses in clinical documents by automatic classification
Context
ICD-9 (International Classification of Diseases, Ninth Revision) is a controlled vocabulary that was published by the WHO in 1977. Based on it, ICD-9-CM (Clinical Modification) is the official system used by United States hospitals in order to assign codes to diagnoses and procedures in the context of routine health care. In 2007, the Cincinnati Computational Medicine Center organized the first automated ICD-9-CM coding challenge. The challenge data set consisted of 1954 anonymized radiology reports collected from the Cincinnati Children's Hospital Medical Center's department of radiology. ICD-9-CM codes were assigned by human coders to each of its records. A total of 45 different codes were used. The challenge data set was then split into the training dataset (978 records) and the application data set (976 records).
Automated coding using a controlled vocabulary (here, the ICD-9-CM classification) is a task which can be performed using term recognition methods as seen above. However, ICD-9-CM class names are often fairly long labels which specify how to classify a disease, rather than actual disease names (for instance, Other specified disorders of kidney and ureter). Therefore, term recognition may not always be appropriate for ICD-9-CM coding.
The challenge was a supervised classification task, since it aimed at learning the human reference codes to be assigned to reports. Furthermore, as it was possible to get more than one reference code for each report, the problem was in fact a multilabel classification task.
Methods
We chose to tackle this multilabeled classification problem using neural networks training. We built a distinct network for each possible ICD-9 code, so that we obtained a family of 45 networks that we finally combined to compute the codes for each record of the application data set.
Evaluation
Detailed quality measures could only be computed on the training data set, as the official code assignment for the application data set was not communicated at the time of the writing of this report. The assembly of networks obtained a precision of 0.95, a recall of 0.90, and an F-measure of 0.92 on the training set. It obtained an F-measure of 0.72 at the official evaluation. The organizers stated that pairwise t-tests using Holm’s correction for multiple comparisons revealed no statistically significant differences between the systems performing at F=0.70 or higher (Pestian et al., 2007).
References
Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Symp. 2001;17-21.
Delbecque T & Zweigenbaum P. MetaCoDe: a lightweight UMLS mapping tool. In Riccardo Bellazzi, Ameen Abu-Hanna, and Jim Hunter, editors, Artificial Intelligence in Medicine, Lecture Notes in Computer Science Volume 4594, Berlin / Heidelberg: Springer, 2007:242-246.
Lindberg DA, Humphreys BL & McCray AT. The Unified Medical Language System. Methods Inf Med, 1993;32(2):81-91.
Pestian JP, Brew C, Matykiewicz P, Hovermale DJ, Johnson N, Cohen KB, Duch W, A shared task involving multi-label classification of clinical free text. In KB Cohen, D Demner-Fushman, C Friedman, L Hirschman, and JP Pestian, editors, ACL 2007 Workshop on Biological, Translational, and Clinical Language Processing, Prague, ACL, 2007:97-104.